
A Game-Changer in Antibiotic Resistance Research
-
by Anoop Singh
- 9
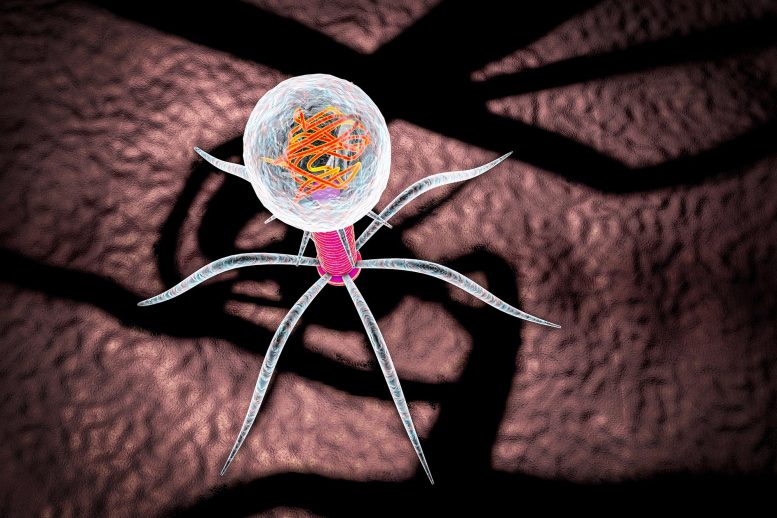
Research on φX174, a bacteriophage, unveils potential in treating antibiotic-resistant infections. Historically significant in phage therapy and molecular biology, φX174’s unique cell-lysing mechanism could pave the way for innovative antibiotics.
φX174, a bacteriophage studied for its potential in combating antibiotic-resistant bacteria, offers new insights into developing alternative antibiotics.
In the age of COVID-19, the word “virus” stirs up thoughts of contagion, sickness, and even death. But what if there were a virus—a very tiny virus capable of replicating itself hundreds of times every half hour—that could cure a severe bacterial infection resistant to all known antibiotics? It is this hope that motivates Bil Clemons, the Arthur and Marian Hanisch Memorial Professor of Biochemistry, to research the virus named φX174.
Understanding φX174
φX174 is a bacteriophage or, more simply, a phage: a virus that targets bacterial cells. From a human perspective, φX174 leads a simple life: It finds its host bacterium, parks on its surface, injects a strand of DNA into the bacterial cell, replicates its DNA over and over again, forces the cell to make viral proteins, assembles the DNA and protein into new virions (copies of the phage), and then breaks open the cell wall of the bacterium so the virions can find other hosts to infect.
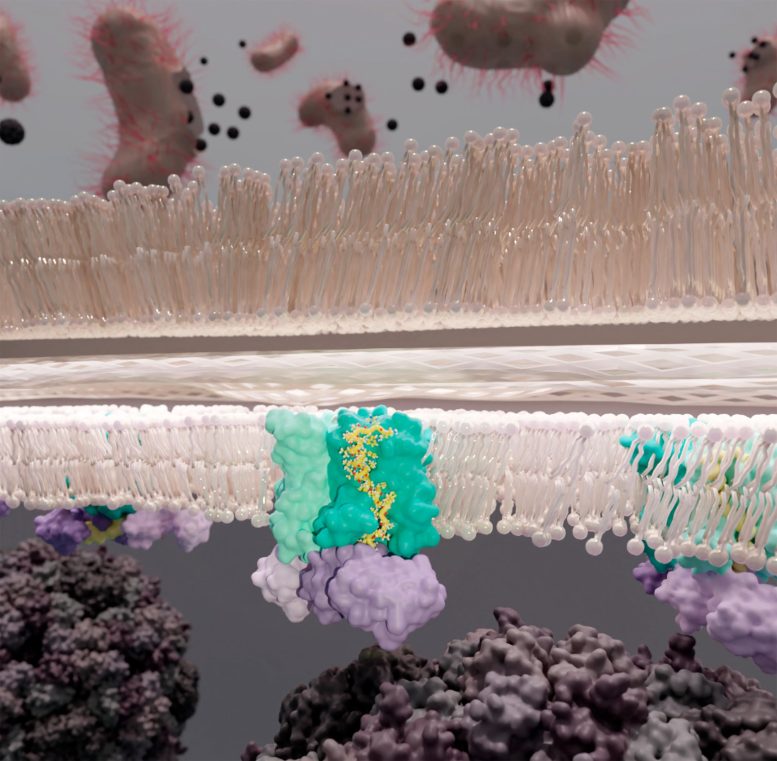
The escape of ΦX174 from its bacterial host. In the membrane is the YES complex (E. coli enzyme MraY [cyan], phage protein E [yellow], & E. coli chaperone SlyD [purple]) where protein E disrupts peptidoglycan synthesis by inhibiting MraY allowing breaching of the cell wall [tan]. Credit: Karen Orta
It is this escape mechanism that the Clemons team elucidates in their paper recently published in the journal Science, “The mechanism of the phage encoded protein antibiotic from φX174.” Relying on images from single-particle electron cryomicroscopy, it is revealed that φX174’s E protein joins with its bacterial host’s proteins MraY and SlyD to form a stable complex—the YES complex. This results in cell lysis: the breaching of the bacterial cell wall and the death of the bacterium.
Historical Context of Phage Discovery
φX174 has been on scientists’ radar for about 100 years. In the early 20th century, the existence of phages was only theorized. Working independently, British bacteriologist Frederick Twort and Quebecois scientist Félix d’Herelle postulated the existence of phages based on the behavior of bacterial cultures in their laboratories. Sometimes, when the bacteria were supposed to be proliferating on their Petri dishes, glossy patches—plaques—would appear where no bacteria grew. Passing these samples through filters captured the bacteria while allowing their tiny invisible killers to go through. Whatever it was that successfully moved through the filters, it was too small to be seen with a microscope.
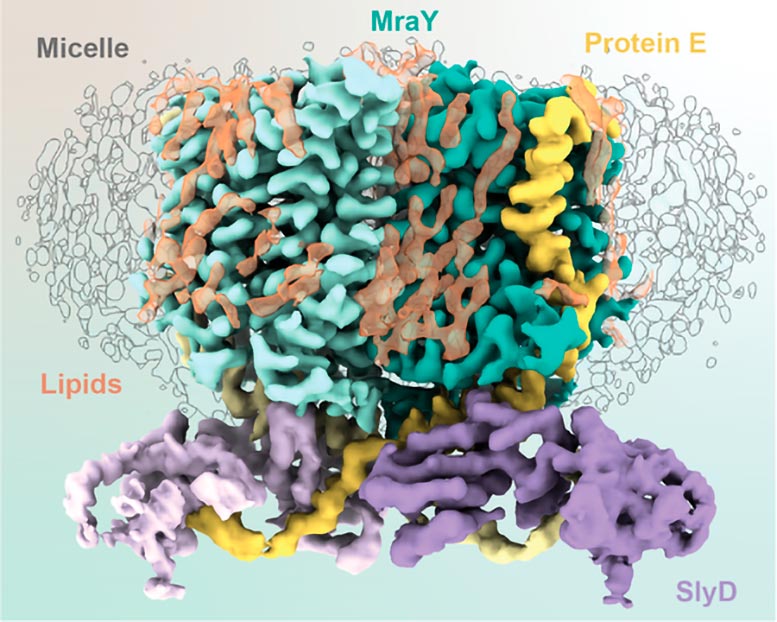
Schematic of the elements comprising the YES complex. Credit: Karen Orta
D’Herelle, working in Paris in 1917, suggested that these killers must be bacteria-eating viruses and was ready to test this theory. According to urban legend, as Clemons relates it, d’Herelle filtered sewage water repeatedly and then drank it to see if it was safe to consume. He felt himself to be unharmed, so he offered a sip to his lab assistant, who was likewise unchanged. D’Herelle then gave the filtered sewage water to a patient, a young boy with severe dysentery who was on the brink of death. With this phage cocktail, which most likely included φX174, the boy was quickly restored to health.
Researchers from across Europe came to Paris to work with d’Herelle. One such researcher, Croatian microbiologist Vladimir Sertič, spent a decade working in d’Herelle’s lab. It was Sertič and his assistant, Nikolai Boulgakov, who devised a taxonomy for known phages. φX174’s exotic sounding name, in Sertic’s classification scheme, simply means “the 174th virus in the tenth [roman numeral X] series of phages that target multiple bacteria,” of the class φ: phages that act against multiple bacteria. Phage therapy continued to cure bacterial diseases, but it killed as well, probably because researchers did not yet know how to purify the byproducts of phage replication such as bacterial debris, which can be toxic.
Evolution of Phage Therapy and Research
Phage research and therapy became fragmented under the pressure of World War II. For the Western allies, the production of highly effective penicillin completely eclipsed phage therapy, becoming the singular solution for bacterial infections. Penicillin was a military secret not shared with the Eastern allies or the Axis powers, so Soviet doctors continued the therapeutic use of phages, a practice that persists today in the countries of the former Soviet Union.
Although phages fell out of favor with medical researchers in Western countries in the decades after World War II, research scientists became fascinated with them. φX174, although only one of billions of different types of phages, moved to the front of the line as a useful experimental tool for the developing field of molecular biology.
Robert L. Sinsheimer, professor of biophysics at Caltech from 1957 to 1977, was instrumental in developing φX174 as a model organism. His lab performed the mapping of φX174’s genome and discovered many of its more intriguing features. As Sinsheimer told the story in a 1991 oral history interview, he invited Max Delbrück, professor of biology at Caltech, to give a series of talks at Iowa State University in the early 1950s where Sinsheimer was then on the faculty. “He [Delbrück] just blew us away with his phage work,” Sinsheimer said. “It was absolutely glorious.”

Max Delbrück with phage group, 1949. Left to right: Jean Jacques Weigle, Ole Maaloe, Elie Wollman, Gunther S. Stent, Max Delbrück, and G. Soli. Credit: Ross Madden, Black Star. 1949-02, 1.3.1-16, Caltech Photographs. California Institute of Technology Archives and Special Collections
Delbrück, who had initially trained as a physicist at the University of Göttingen before the war, was building a cadre of phage researchers at Caltech and using the viruses to probe the mysteries of molecular genetics. Sinsheimer made it his mission to come to Caltech during a six-month leave in 1953 to learn how to work with phages. One day while sitting in Delbrück’s office discussing how to proceed with virology, the two men concluded that it might be beneficial to study the smallest and potentially simplest phages to better understand viral structure and replication. Sinsheimer reviewed phage candidates, settled on φX174, acquired samples from labs in England and France, and set to work.
There began a string of firsts for science based on φX174. In a 1966 essay, Sinsheimer referred to φX174 as “multum in parvo”: Latin for “much in little.” Throughout the 1950s and 1960s, φX174 continually surprised researchers. In 1959, two years after joining Caltech, Sinsheimer determined that φX174 had only a single strand of DNA that it injected into the host cell to begin replication. This was a surprise given that DNA had been discovered to have a double-helical structure only a few years earlier. In 1962, Sinsheimer speculated that φX174’s DNA was shaped like a circular ring, something molecular biologists had not yet visualized. In 1977, Frederick Sanger of the University of Cambridge was the first person to completely sequence a genome, earning him the 1980 Nobel Prize in Chemistry. That genome belonged to φX174. The phage itself was acquired from Sinsheimer.

Robert Sinsheimer in his laboratory, 1974. Credit: Floyd Clark. 10.24-116, Caltech Photographs. California Institute of Technology Archives and Special Collections
By the late 1970s, much of the life cycle of φX174 was well understood, but uncertainties remained. It was assumed that φX174 broke out of its bacterial host by blocking the synthesis of the peptidoglycan layer—a key protective barrier in the cell wall of all bacteria—just as penicillin and other pharmaceutical antibiotics do.
For most phages, scientists had learned how they make specialized enzymes, endolysins, that degrade the sugar–amino acid polymer that makes up the peptidoglycan layer. But these enzymes are too large to be contained in the DNA of a tiny phage like φX174.
Modern Research on φX174
“The φX174 genome is really small,” Clemons explains. “If you were to encode something that achieves cell lysis in the way a lysozyme does—an enzyme found in our tears and saliva that provide protection against bacteria by mimicking endolysins—there would be no room for other proteins on the φX174 genome. φX174 is part of a group of these viruses that are too small to have complex lysis machinery, so these phages had to evolve very simple ways of lysing bacterial cells.”
Different phages and antibiotics interfere with the synthesis of peptidoglycan at varying points in the process. The E protein of φX174 targets MraY, a membrane enzyme that catalyzes the synthesis of a peptidoglycan precursor. To complete its destructive work, φX174’s protein E requires another protein, SlyD, which it hijacks from its bacterial host. “It’s a mystery,” says Clemons, “because SlyD has no reason to act here. It normally does not interact with MraY, it has an entirely different job. Yet somehow, this process requires SlyD.”
These three agents, one viral and two from the host, comprise the YES complex: MraY, protein E, SlyD. Essentially, the E protein of φX174 entwines itself with MraY, inhibiting MraY’s enzymatic activity. SlyD binds and stabilizes the protein E and MraY complex without contacting MraY.
Implications for Future Antibiotic Development
This discovery stands poised to help researchers fulfill bacteriophages’ initial promise as an antibiotic therapeutic. Antibiotics have saved countless numbers of lives over the past century, but the invention of new classes of antibiotics has been unable to keep up with the ability of bacteria to develop resistance to them. Bacteria also mutate to resist phages, but unlike pharmaceutical antibiotics that require extensive human effort to improve their structure, phages themselves can mutate, countering new bacterial defenses. We live with a tremendous number of phages in our bodies, many hundreds of trillions. It is the hope of Clemons and other researchers in the field that marshaling the right phages at the right time to address bacterial infections could create a new, more durable antibiotic, one we increasingly need as we confront antibiotic resistant bacteria.
Reference: “The mechanism of the phage-encoded protein antibiotic from ΦX174” by Anna K. Orta, Nadia Riera, Yancheng E. Li, Shiho Tanaka, Hyun Gi Yun, Lada Klaic and William M. ClemonsJr., 14 July 2023, Science.
DOI: 10.1126/science.adg9091
Co-authors of “The mechanism of the phage encoded protein antibiotic from φX174” include Anna K. Orta, Nadia Riera, Evelyn Yancheng Li, Shiho Tanaka, Hyun Gi Yun, and Lada Klaic. Funding was provided by the National Institutes of Health and the G. Harold and Leila Y. Mathers Foundation.
Research on φX174, a bacteriophage, unveils potential in treating antibiotic-resistant infections. Historically significant in phage therapy and molecular biology, φX174’s unique cell-lysing mechanism could pave the way for innovative antibiotics. φX174, a bacteriophage studied for its potential in combating antibiotic-resistant bacteria, offers new insights into developing alternative antibiotics. In the age of COVID-19, the word…
Research on φX174, a bacteriophage, unveils potential in treating antibiotic-resistant infections. Historically significant in phage therapy and molecular biology, φX174’s unique cell-lysing mechanism could pave the way for innovative antibiotics. φX174, a bacteriophage studied for its potential in combating antibiotic-resistant bacteria, offers new insights into developing alternative antibiotics. In the age of COVID-19, the word…
